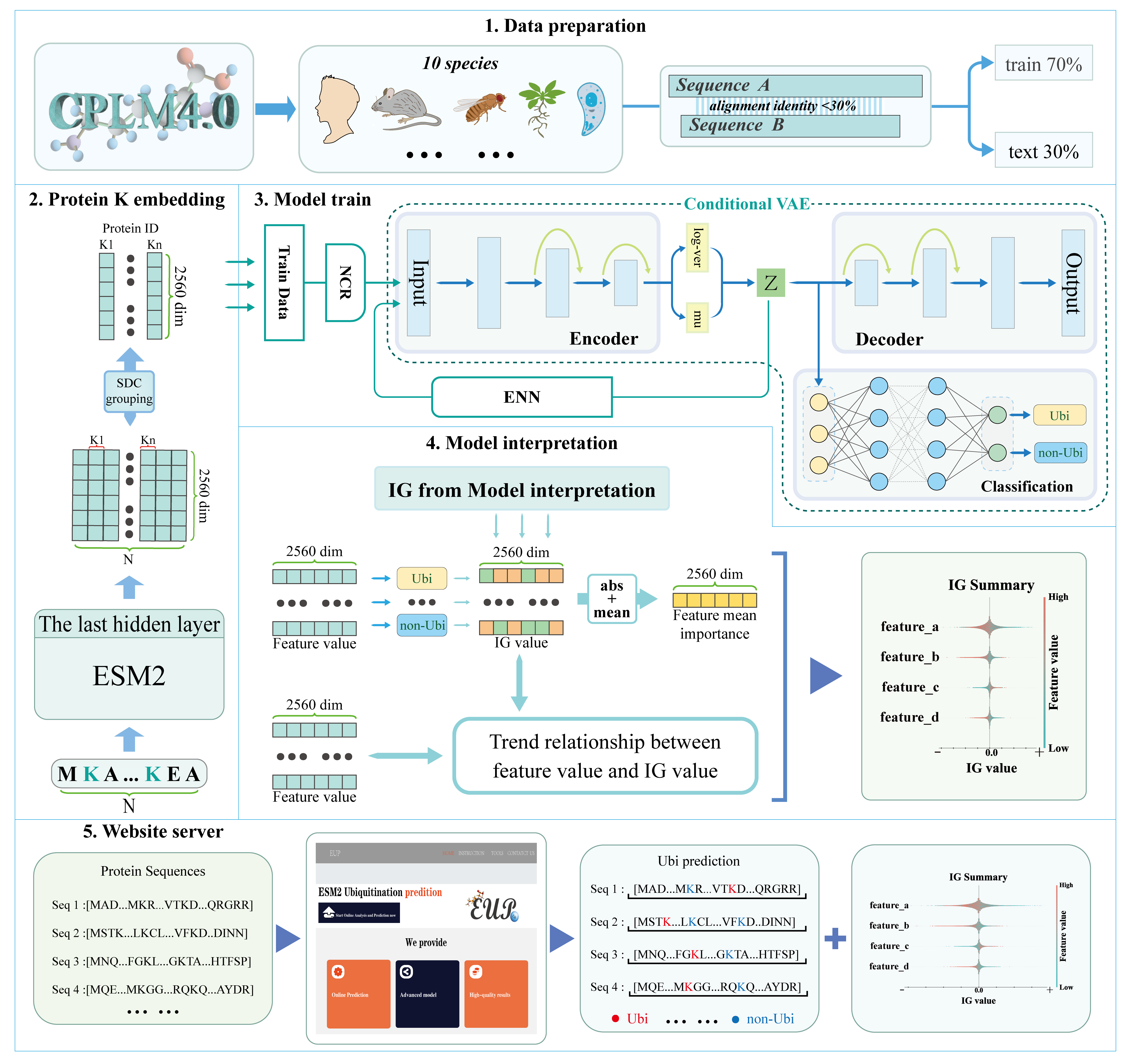
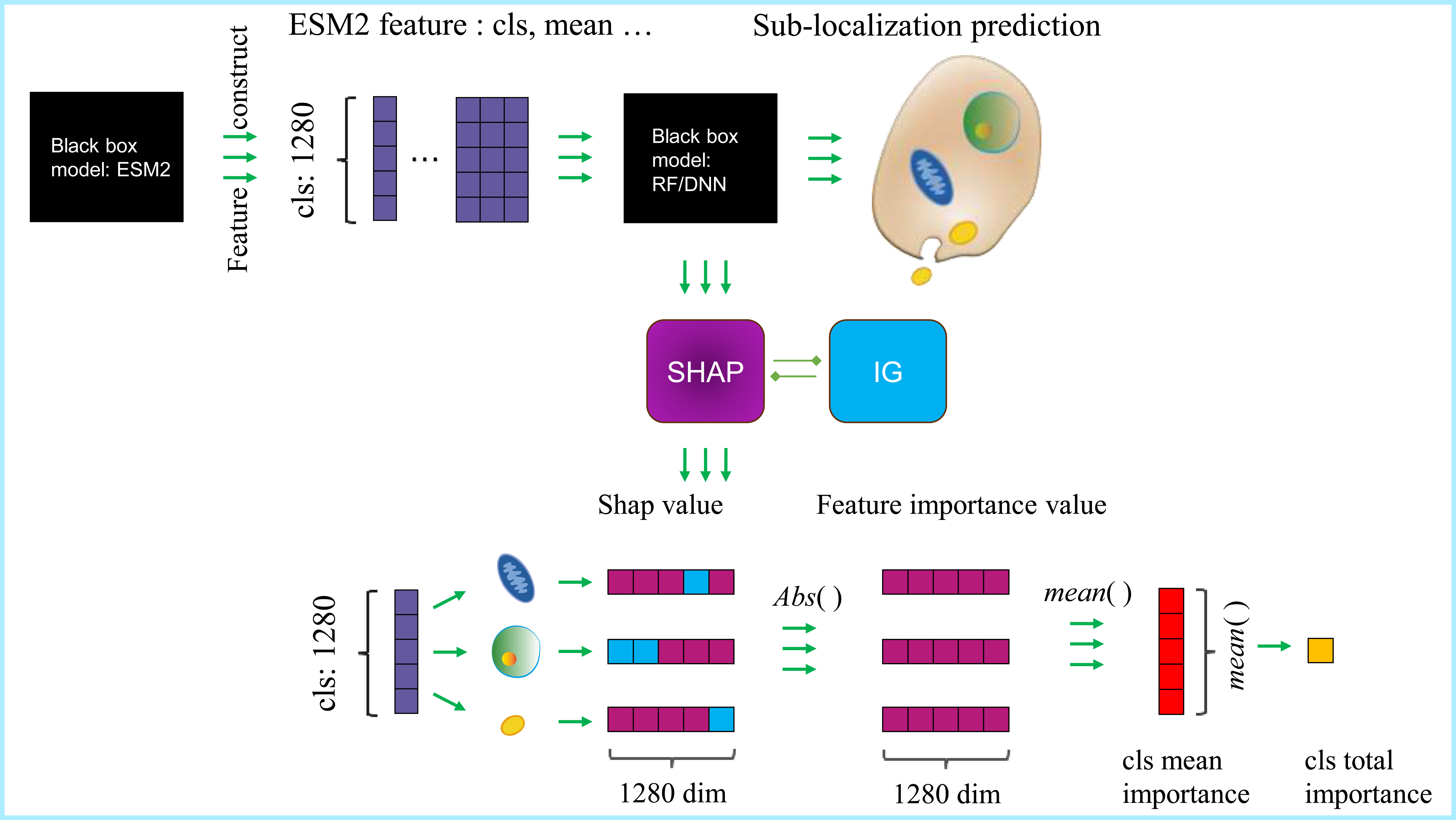
brief description of this work
How to use online analysis tools
This online analysis model is based on the feature representation of lysine sites extracted from ESM2, which has been used to construct a cVAE_ResDNN model for ubi-prediction. Users can submit protein sequences to this online prediction tool, and can get the ubi-prediction outcomes.
1. Using the Online Upload Analyze Service (for ubi-prediction)
Step 1: Users need to log in to "https://eup.aibtit.com/upload" to use this service.
Step 2: Users need to provide the protein sequence in FASTA format for testing (for example data, please see the "Download Example Data" on this page).
Step 3: After submitting, users will receive an extraction code and can check the processing status of their test file.
Step 4: Once the processing is complete with the extraction code, users can download the results in an Excel spreadsheet and visualization images of ubiquitination sites.
2. Using the SHAP Visualization Service (for model interpretation)
Step 1: Users need to log in to "https://eup.aibtit.com/shap_upload" to use this service.
Step 2: Users need to upload the result Excel file from the Online Upload Analyze service (containing 2560 features) in the required format (for example data, please see the "Download Example Data" on this page).
Step 3: After submitting, users will receive an extraction code and can check the processing status of their test file.
Step 4: Once the processing is complete with the extraction code, users can download the SHAP visualization images for feature interpretation and further analysis.